4. Kymographs¶
Download this page as a Jupyter notebook
We can download the data needed for this tutorial directly from Zenodo using Pylake.
Since we don’t want it in our working folder, we’ll put it in a folder called "test_data":
filenames = lk.download_from_doi("10.5281/zenodo.7729525", "test_data")
We can use the lk.File.kymos attribute to access the kymographs from a file:
import lumicks.pylake as lk
file = lk.File("test_data/kymo.h5")
print(file.kymos) # dict of available kymos: {'16': Kymo(pixels=699)}
This is a regular Python dictionary so we can easily iterate over it:
# print some details for all kymos in a file
for name, kymo in file.kymos.items():
print(f"kymograph '{name}', starts at {kymo.start} ns")
# kymograph '16', starts at 1638534513847557200 ns
Or access a particular Kymo object directly to work with:
# use the kymo name as a dict key
kymo = file.kymos["16"]
4.1. Plotting and exporting¶
Pylake provides a convenience plot() method to quickly
visualize your data. For details and examples see the Plotting Images section.
The kymograph can also be exported to TIFF format:
kymo.export_tiff("image.tiff")
4.2. Kymo data and details¶
We can access the raw image data as a numpy.ndarray:
rgb = kymo.get_image("rgb") # matrix with `shape == (height, width, 3 colors)`
blue = kymo.get_image("blue") # single color so `shape == (height, width)`
# Plot manually
plt.figure()
plt.imshow(kymo.get_image("green"), aspect="auto", vmax=15)
plt.show()
There are also several properties available for convenient access to the kymograph metadata:
kymo.center_point_umprovides a dictionary of the central x, y, and z coordinates of the scan in micrometers relative to the brightfield field of viewkymo.size_umprovides a list of scan sizes in micrometers along the axes of the scankymo.pixelsize_umprovides the pixel size in micrometerskymo.pixels_per_lineprovides the number of pixels in each line of the kymographkymo.fast_axisprovides the axis that was scanned (x or y)kymo.line_time_secondsprovides the time between successive lineskymo.pixel_time_secondsprovides the pixel dwell time.kymo.durationprovides the full duration of the kymograph in seconds. This is equivalent to the number of scan lines timesline_time_seconds.
4.3. Slicing, cropping & flipping¶
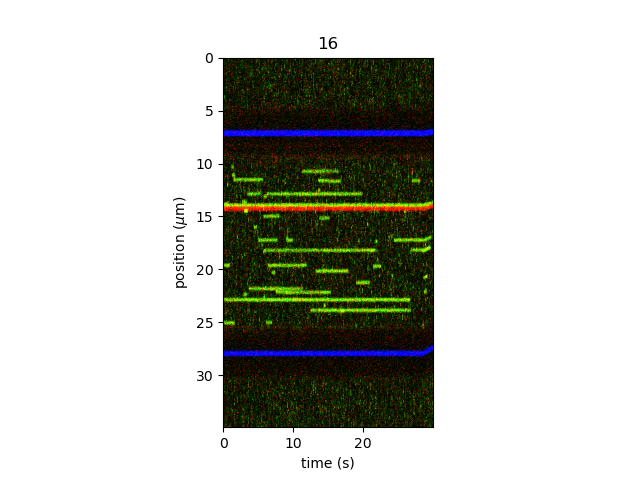
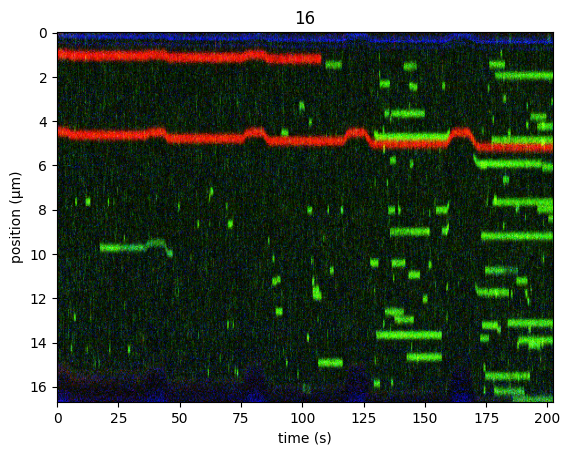
Kymographs can be sliced in order to obtain a specific time range. For example, one can plot the region of the kymograph between 130 and 160 seconds using:
plt.figure()
kymo["130s":"160s"].plot("rgb", adjustment=lk.ColorAdjustment(0, 98, mode="percentile"))
plt.show()
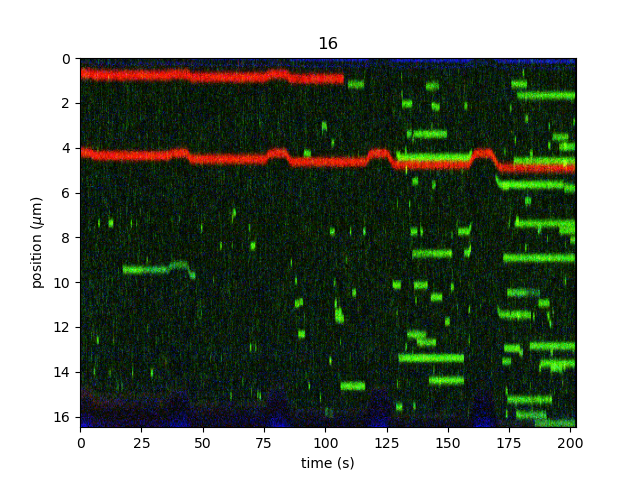
It is possible to crop a kymograph to a specific coordinate range, by using the function
Kymo.crop_by_distance(). For example, we can
crop the region from 9.5 micron to 26 microns using the following command:
plt.figure()
kymo.crop_by_distance(9.5, 26).plot("rgb", aspect="auto", adjustment=lk.ColorAdjustment(0, 98, mode="percentile"))
plt.show()
If we know the bead diameter, we can automatically crop the kymo to an estimate of the bead edges using crop_beads().
This can be convenient when batch processing many kymographs:
plt.figure()
kymo.crop_beads(4.84, algorithm="brightness").plot("rgb", aspect="auto", adjustment=lk.ColorAdjustment(0, 98, mode="percentile"))
plt.show()
Note
Note, slicing in time is currently only supported for unprocessed kymographs. If you want to both crop and slice a kymo, the order of operations is important – you need to slice before cropping:
kymo_sliced = kymo["130s":"160s"]
kymo_cropped = kymo_sliced.crop_by_distance(9.5, 26)
plt.figure()
kymo_cropped.plot("rgb", adjustment=lk.ColorAdjustment(0, 99.9, mode="percentile"))
plt.show()
If you try to slice a kymograph that has already been cropped, a NotImplementedError will be raised.
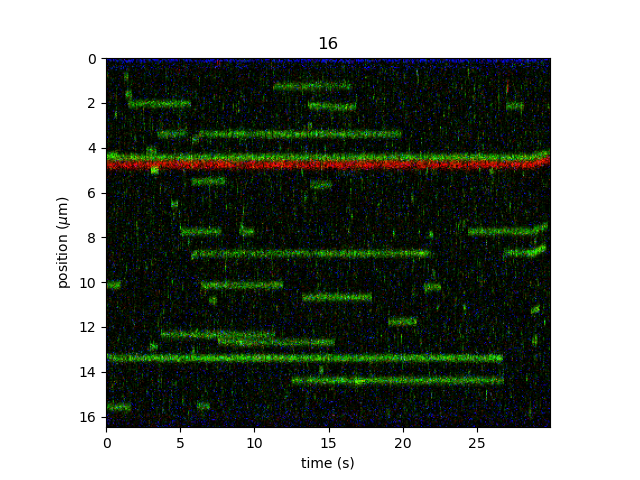
Finally, we can also flip a kymograph along its positional axis using flip().
This returns a new (but flipped) Kymo:
kymo_flipped = kymo.flip()
plt.figure()
plt.subplot(211)
kymo.plot("rgb", adjustment=lk.ColorAdjustment(0, 98, mode="percentile"))
plt.subplot(212)
kymo_flipped.plot("rgb", adjustment=lk.ColorAdjustment(0, 98, mode="percentile"))
plt.tight_layout()
plt.show()
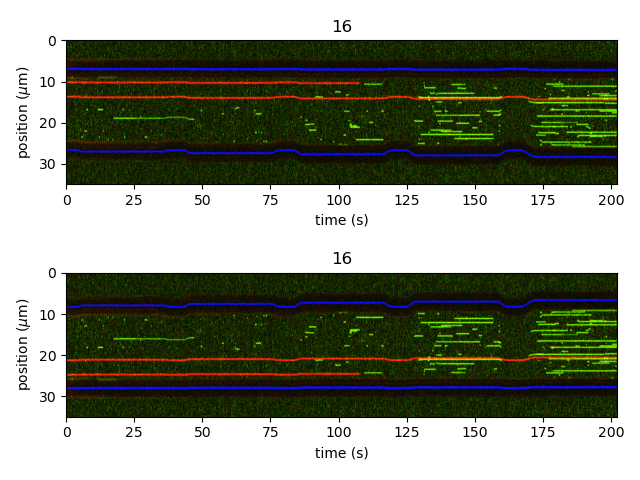
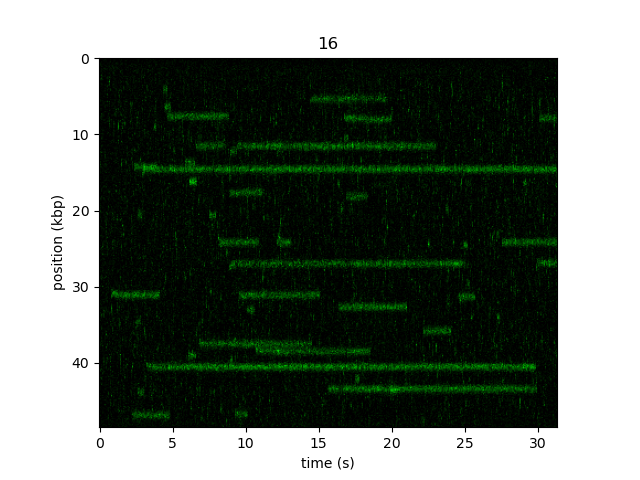
4.4. Calibrating to base pairs¶
By default, kymographs are constructed with units of microns for the position axis. If, however, the kymograph spans a known length of DNA (here for example, lambda DNA) we can calibrate the position axis to kilobase pairs (kbp):
kymo_kbp = kymo_cropped.calibrate_to_kbp(48.502)
Now if we plot the image, the y-axis will be labeled in kbp:
plt.figure()
kymo_kbp.plot("green")
plt.show()
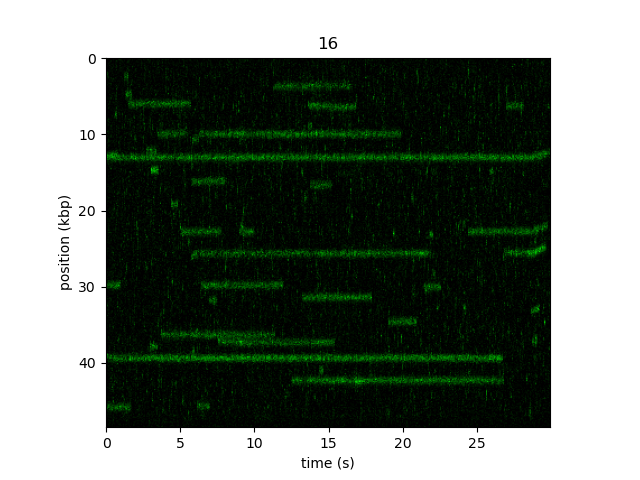
These units are also carried forward to any downstream operations such as kymotracking algorithms and MSD analysis.
Warning
Currently this is a static calibration, meaning it is only valid if the traps do not change position during the time of the kymograph.
Also, the accuracy of the calibration is dependent on how the kymo is cropped. If you crop the kymo by visually estimating the bead edges, the resulting position should be taken as approximate.
4.5. Interactive slicing, cropping & calibration¶
We can also interactively slice, crop, and calibrate kymographs using crop_and_calibrate():
widget = kymo.crop_and_calibrate(channel="rgb", tether_length_kbp=48.502, aspect="auto", adjustment=lk.ColorAdjustment(0, 99.5, mode="percentile"))
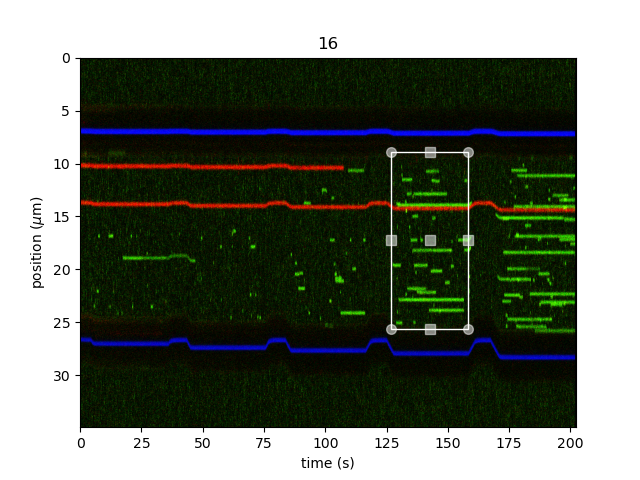
Simply click and drag the rectangle selector to the desired ROI. We can then access the edited kymograph with:
new_kymo = widget.kymo
plt.figure()
new_kymo.plot("green")
plt.show()
If the optional tether_length_kbp argument is supplied, the kymograph is automatically calibrated to the desired
length in kilobase pairs. If this argument is missing (the default value None) the edited kymograph is only
sliced and cropped.
4.6. Downsampling¶
We can downsample a kymograph in time by invoking:
kymo_ds_time = kymo_cropped.downsampled_by(time_factor=2)
Or in space by invoking:
kymo_ds_position = kymo_cropped.downsampled_by(position_factor=2)
Or both:
kymo_ds = kymo_cropped.downsampled_by(time_factor=2, position_factor=2)
adjustment = lk.ColorAdjustment(0, 30, mode="absolute")
plt.figure()
plt.subplot(221)
kymo_cropped.plot("green", adjustment=adjustment)
plt.title("original")
plt.subplot(222)
kymo_ds_time.plot("green", adjustment=adjustment)
plt.title("downsampled time")
plt.subplot(223)
kymo_ds_position.plot("green", adjustment=adjustment)
plt.title("downsampled position")
plt.subplot(224)
kymo_ds.plot("green", adjustment=adjustment)
plt.title("downsampled both")
plt.tight_layout()
plt.show()
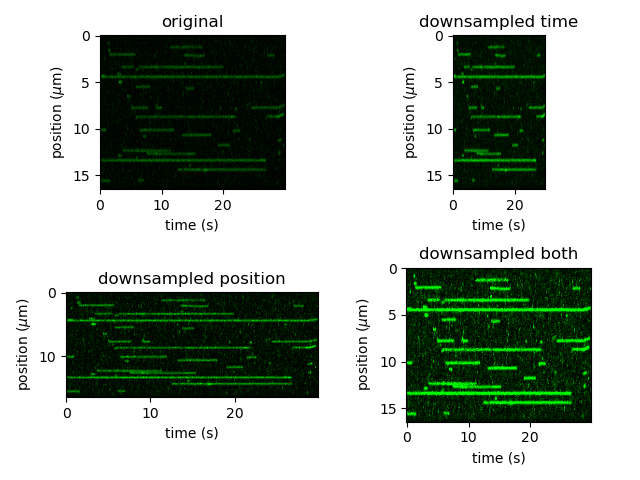
Note however, that not all functionalities are present anymore when downsampling a kymograph over time. This is because the downsampling occurs over non-contiguous sections of time (across multiple scan lines) and therefore each pixel no longer has an identifiable time. For example, we can no longer access the per pixel timestamps:
# this cell will raise a `NotImplementedError`
kymo_ds.timestamps
Additionally, a downsampled kymograph cannot be sliced (same as cropped kymographs mentioned above). Therefore you should first slice the kymograph and then downsample.
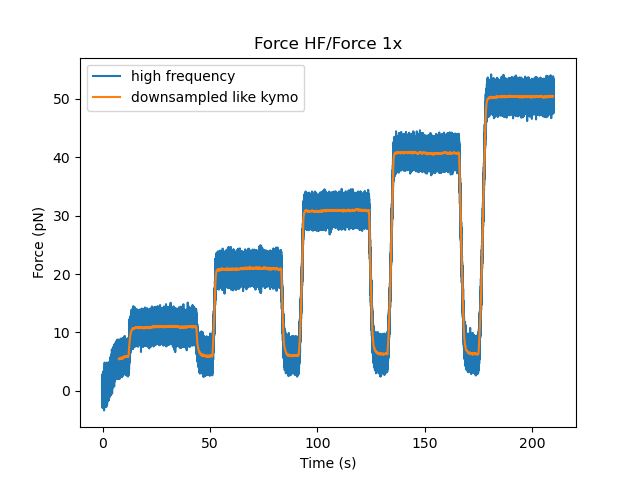
4.7. Correlating with force¶
We can downsample channel data according to the lines in a kymo. We can use
line_timestamp_ranges() for this:
line_timestamp_ranges = kymo.line_timestamp_ranges()
This returns a list of start and stop timestamps that can be passed directly to downsampled_to(),
which will then return a Slice with a datapoint per line:
force = file.force1x
downsampled = force.downsampled_over(line_timestamp_ranges)
plt.figure()
force.plot(label="high frequency")
downsampled.plot(start=force.start, label="downsampled like kymo")
plt.legend()
plt.show()
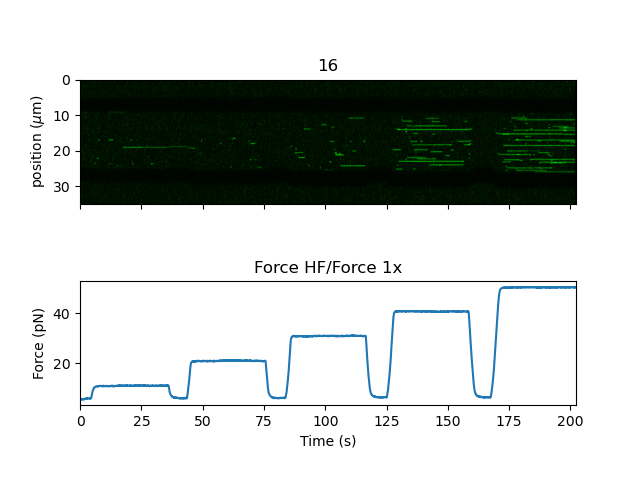
There is also a convenience function plot_with_force() to plot a kymograph along with a
downsampled force trace:
kymo.plot_with_force("1x", "green", adjustment=lk.ColorAdjustment(0, 15))
This will average the forces over each Kymograph line and plot them in a correlated fashion.
The function can also take a dictionary of extra arguments to customize the kymograph plot.
These parameter values get forwarded to matplotlib.pyplot.imshow().